What: Analysing data with a data workflow, fast Java startup & csv Magic
Why: Building data analysis pipelines for (small) problems, where intermediate steps are automatically documented
How: Use Drake for (data) workflow management, Drip for fast JVM startup and csvkit for csv magic.
In this post, I will show you how to build a small data analysis pipeline for analysing the references of a wikipedia article (about data science). The result is the following simple image, all steps are automated and intermediate results are documented automatically.
In the end, you should have four artifacts documenting your work:
- Drakefile: The workflow file, with which you ca regenerate all other artifacts. Plain text, thus can be easily used with version control systems.
- data.collect: The html content of the wikipedia article as the source of the analysis
- data.extract: The publishing years of the references with number of occurrence
- result.plot.png: A png of the publishing year histogram
Agenda
- Install the requirements
- Build the pipeline
- Run the pipeline
Install the requirements
You need the following tools:
- Linux (Debian or Ubuntu, command line tools)
- Python (for html processing and csv magic)
- R (for plotting)
- Java (for Drake)
You can install the dependencies easily with the script below. The following steps are tested within a Debian (Jessie) VM, 64bit. It should also work on Ubuntu. Maybe, other distros have to be adapted.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 | # Update sudo apt-get update # Install curl sudo apt-get install -y curl # Install R sudo apt-get install -y r-base sudo apt-get install -y r-cran-rcpp sudo Rscript -e 'install.packages("ggplot2", repos="https://cran.uni-muenster.de/")' # Install Java8 sudo sh -c 'echo deb http://ftp.de.debian.org/debian jessie-backports main >> /etc/apt/sources.list' sudo apt-get update sudo apt-get install -y openjdk-8-jdk # Install Drip mkdir ~/.lib git clone https://github.com/flatland/drip.git ~/.lib/drip cd ~/.lib/drip make prefix=~/.bin install # Download & Setup Drake wget https://github.com/Factual/drake/releases/download/1.0.3/drake.jar -O ~/.lib/drake.jar cat << 'EOF' > ~/.bin/drake #!/bin/bash drip -cp ~/.lib/drake.jar drake.core "$@" EOF chmod u+x ~/.bin/drake echo export PATH=~/.bin:$PATH >> ~/.bashrc # Install csvkit pip install csvkit |
Build the pipeline
Drake is controlled by a so called Drakefile. Let us define three steps for the data processing:
- Data collection (html from Wikipedia)
- Data extraction (Extracting the reference texts and years of the references)
- Plotting results (Plotting the results to png)
1. Data collection
The first step can be done with Linux internal tools. Thus, we can create the Drakefile with the first step already:
data.collect <- [-timecheck] curl -k https://de.wikipedia.org/wiki/Data_Science > $OUTPUT
Drake takes input (mostly) from files and sends the output of each step again to files. Thus, the result of each step in a workflow is automatically documented.
This first step will download the html for the data science article from the german wikipedia and stores the html in a file called data.collect (the [-timecheck] avoids running the step each time drake is started because of missing input from previous steps). You can already run this workflow with the drake command:
drake |
This will generate a file called data.collect containing the html of the wikipedia page.
2. Data extraction
For data extraction from html, Python & BeautifulSoup is used. The extraction of the year from each reference can be done with linux internal tools (for example grep). Thus, the python program should read from stdin, get the reference text and outputs plain text to stdout. Create a file called extract.py with the following content:
1 2 3 4 5 6 7 8 9 10 11 12 | #!/usr/bin/env python3 from bs4 import BeautifulSoup import sys input=sys.stdin.read() soup=BeautifulSoup(input, 'html.parser') entries=soup.find('ol', {'class': 'references'}).findAll('li') for entry in entries: print(entry.getText()) |
Make the script file executable with:
chmod u+x ./extract.py |
You can test the script with the following command (If you ran the workflow from step 1 before):
cat data.collect | ./extract.py |
Now, let us extend the Drakefile to use the python script, search for years with a regex, create a histogram by counting the occurrences of the years, reorder the columns and add a header:
data.collect <- [-timecheck]
curl -k https://de.wikipedia.org/wiki/Data_Science > $OUTPUT
data.extract <- data.collect
cat $INPUT | ./extract.py | grep -o -P '\b\d{4}\b' | sort | uniq -c | sort -nr | \
sed 's/^[ ]*//g' | csvcut -c2,1 -d " " | printf 'year, occ'"\n$(cat)" > $OUTPUT
If you run this workflow with:
drake |
a new file called data.extract will be created which looks like the following:
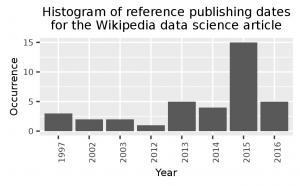
year, occ 2015,15 2016,5 2013,5 2014,4 1997,3 2003,2 2002,2 2012,1 1145,1
Please note the wrongly detected date 1145.
You can filter stupid dates out with csvsql (I really recommend this tool) from the csvkit tool suite by extending the Drakefile with some simple sql:
data.collect <- [-timecheck]
curl -k https://de.wikipedia.org/wiki/Data_Science > $OUTPUT
data.extract <- data.collect
cat $INPUT | ./extract.py | grep -o -P '\b\d{4}\b' | sort | uniq -c | sort -nr | \
sed 's/^[ ]*//g' | csvcut -c2,1 -d " " | printf 'year, occ'"\n$(cat)" | \
csvsql --query 'select * from stdin where year>1900 and year<2100' > $OUTPUT
Plotting results
The final step is the plotting of the results. Let us create a R file, which reads from stdin and plots data to png. Create the file plot.R with the following content:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 | #!/usr/bin/env Rscript
library(ggplot2)
data <- read.table(pipe('cat /dev/stdin'), header=T, sep=",")
data[,1]<-factor(data[,1])
names<-names(data)
p<-ggplot(data, aes(x=data[, 1], y=data[, 2]))+geom_bar(stat='identity')+
theme(axis.text.x = element_text(angle = 90, hjust = 1))+
xlab(names[1])+ylab(names[2])+theme(text = element_text(size=8))
args = commandArgs(trailingOnly=TRUE)
if(length(args)>1){
t<-paste(strwrap(args[2], width=40), collapse = "\n")
p+ggtitle(t)
}
ggsave(args[1], width = 2*1.618, height = 2) |
Make the script file executable with:
chmod u+x ./plot.R |
Now extend the Drakefile with the last step: Image creation. If Drake is run again, it is checking if the output of steps is already up to date by checking for files with the step name. Thus, the image file name and the step should match:
data.collect <- [-timecheck]
curl -k https://de.wikipedia.org/wiki/Data_Science > $OUTPUT
data.extract <- data.collect
cat $INPUT | ./extract.py | grep -o -P '\b\d{4}\b' | sort | uniq -c | sort -nr | \
sed 's/^[ ]*//g' | csvcut -c2,1 -d " " | printf 'year, occ'"\n$(cat)" | \
csvsql --query 'select * from stdin where year>1900 and year<2100' > $OUTPUT
result.plot <- data.extract
cat $INPUT | ./plot.R $OUTPUT \
"Histogram of reference publishing dates for the Wikipedia data science article"
Again, run this step by executing the drake command.
Run the pipeline
Finally, to see all steps working together delete the generated artifacts (data.collect, data.extract, result.plot.png) and run drake again:
vagrant@debian-jessie:/tmp$ drake The following steps will be run, in order: 1: /tmp/././data.collect <- [missing output] 2: /tmp/././data.extract <- /tmp/././data.collect [projected timestamped] 3: /tmp/././result.plot.png <- /tmp/././data.extract [projected timestamped] Confirm? [y/n] y Running 3 steps with concurrence of 1... --- 0. Running (missing output): /tmp/././data.collect <- % Total % Received % Xferd Average Speed Time Time Time Current Dload Upload Total Spent Left Speed 100 58980 0 58980 0 0 172k 0 --:--:-- --:--:-- --:--:-- 196k --- 0: /tmp/././data.collect <- -> done in 0.54s --- 1. Running (missing output): /tmp/././data.extract <- /tmp/././data.collect --- 1: /tmp/././data.extract <- /tmp/././data.collect -> done in 0.28s --- 2. Running (missing output): /tmp/././result.plot.png <- /tmp/././data.extract --- 2: /tmp/././result.plot.png <- /tmp/././data.extract -> done in 0.75s Done (3 steps run). |
You should now have three files for each of the workflow steps, where the last step file contains the image shown above.